by Pankaj Bhatt ,†,Amit Verma ,*,†,Shulbhi Verma ,Md. Shahbaz Anwar ,Parteek Prasher ,Harish Mudila and Shaohua Chen*
* Authors to whom correspondence should be addressed.
† Both authors contributed equally to this manuscript.
Keywords: phytomicrobiome; rhizosphere engineering; organic contamination; microorganisms
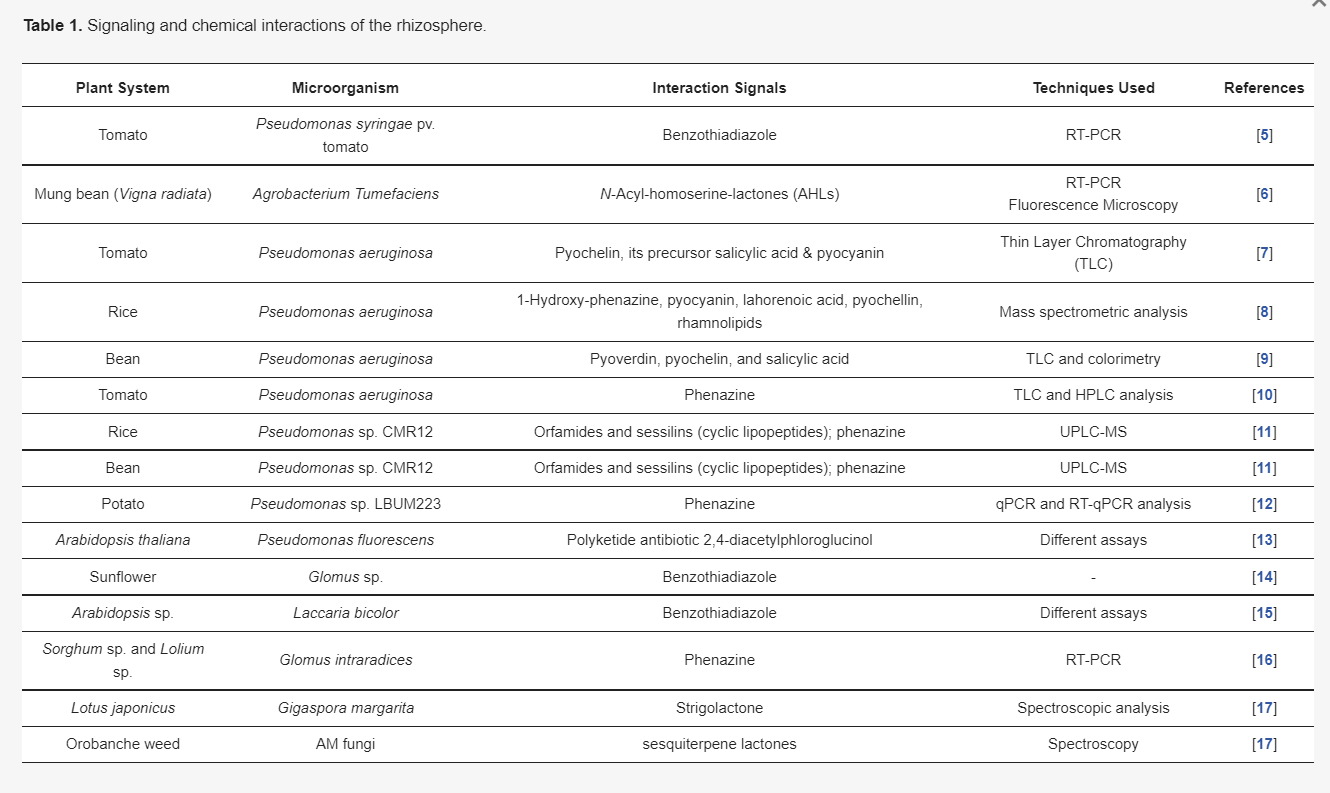
Table 1. Signaling and chemical interactions of the rhizosphere.
The microbial community interacts with the plant not only in the rhizospheric region but also in the phyllosphere, which contains a wide array of living organisms such as bacteria, fungi, yeasts, algae, nematodes, etc. It influences the phytophysiology and overall development [18]. The biotic and abiotic conditions regulate the microbiome composition of the phyllosphere [19]. This PM also varies with biotic factors, such as a pathogenic organism causing disease, where the host plant microbiome varies with disease progression [20]. Thus, understanding the phyllosphere microbial community gives an informative overview of plant health. PM analysis requires a holistic study involving the different techniques of genomics, transcriptomics, proteomics, and metabolomics to determine the relationship between the plant and its associated microbial community. An amplified rDNA restriction analysis (ARDRA) is usually utilized for characterizing the bacterial community present in any given sample, which in short involves enzymatic amplification followed by the restriction digestion of 16S rRNA amplified fragments [21]. Advanced techniques like Illumina-based microbiome analysis further helps to analyze the microbial community and their dynamic relationships in much less time [20,22]. Thus, recently, much research has been carried out concerning the PM structure and composition and their variations under different plant conditions.
Figure 1. Factors regulating the microbiome composition.
Studies related to PM compositions have revealed that plants are in dynamic relationships with the surrounding microbial communities, which show variations as per the plant developmental stage and their environmental conditions. Either the rhizosphere or phyllosphere or even the endophytic microbes are seen to enable plant establishment in their surroundings. It is very interesting to note that the same plant grown in different soils shows the recruitment of the same microbial communities for their establishment, which strongly reflects the modifications of the soil microbiota by plants. This underlies an intricate pathway of signaling compounds for community assembly, which opens a new avenue of research. The root microbiome of the plant of different species shows variations in community structure, along with the presence of Actinobacteria, Proteobacteria, and Bacteriodetes as conserved phyla in the rhizomicrobiome of these plant species [27]. The plants Arabidopsis thaliana, A. lyrata, A. helleri, and Cardamine hirsute, which were grown in the same soil in the mentioned study, reflect the requirement for the exploration of PM signaling for sustainable cultivation.
Similarly, in phyllosphere studies it has been observed by many workers that there is little variation in the phyllomicrobiome among the similar plant species, even those planted in various geographical areas [28,29]. Recently, these PM-associated microorganisms were found to be crucial in imparting tolerance against biotic and abiotic stress. The role of the rhizosphere communities is well established, but in recent years workers have reported the involvement of various phyllospheric communities in plant stress alleviation. The phyllosphere bacteria of the Bacillus genus have been reported to mitigate drought stress in rice by previous researchers [30]. The phyllosphere isolates were found to be involved in the activation of the plant antioxidant system by an increase in proline-like osmolytes; phenolic content; and the activity of antioxidative enzymes, such as superoxide dismutase, catalase, peroxidase, etc. [31]. Such studies are now easy to conduct with the advent of what we call “next-generation sequencing”, which includes the methods of 454 pyrosequencing, Illumina sequencing, etc. As compared to the traditional techniques, these tools are advantageous and can be used for multidirectional analysis. Further, these techniques are supplemented by metaproteomic analysis, revealing the complex networks of these PM [32]. Thus, we can say that understanding the ecology of the PM is presently an important thrust area of research to achieve sustainability in agriculture in the coming years. Some of the recent PM studies are presented in Table 2.
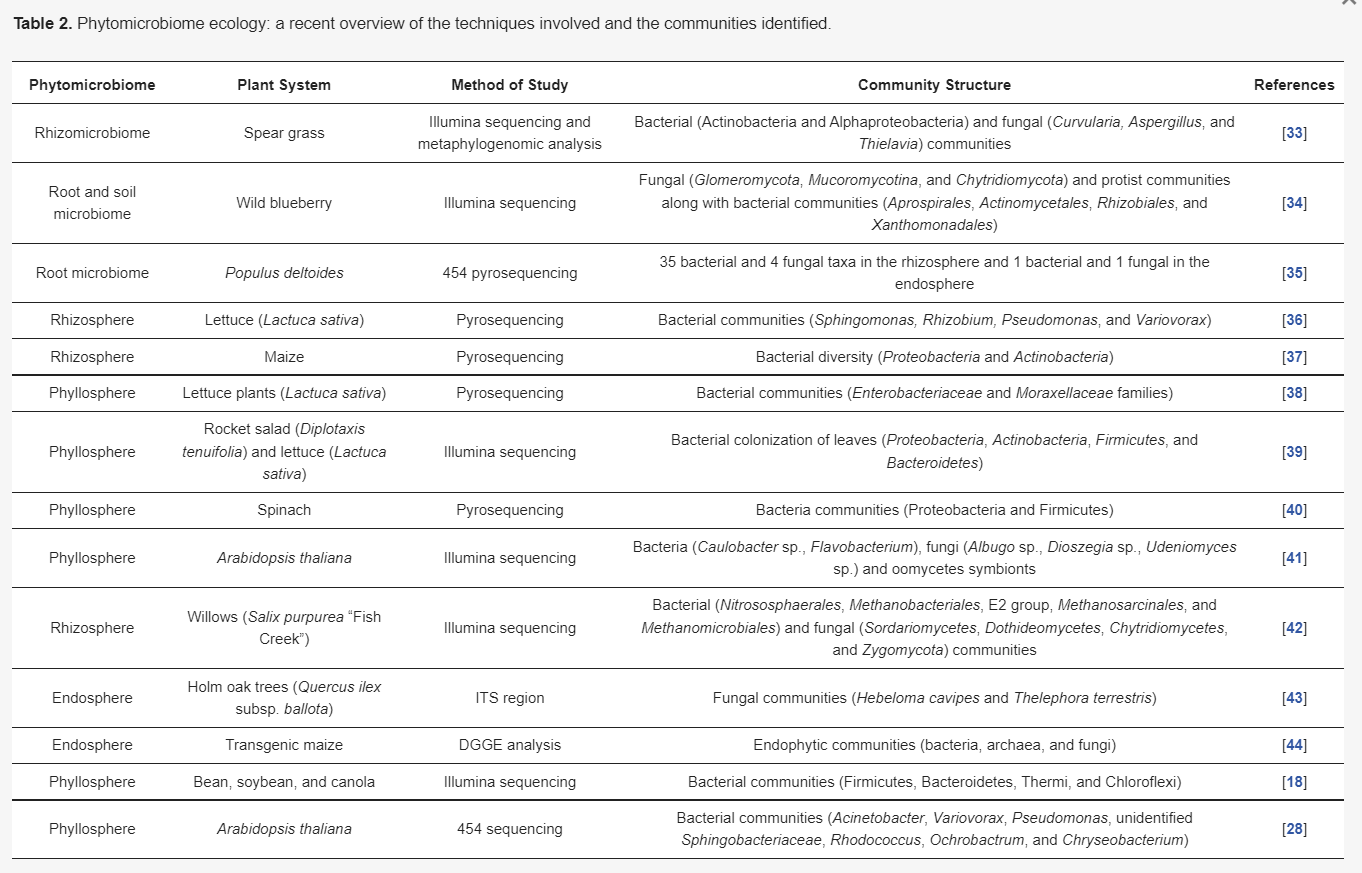
Table 2. Phytomicrobiome ecology: a recent overview of the techniques involved and the communities identified.
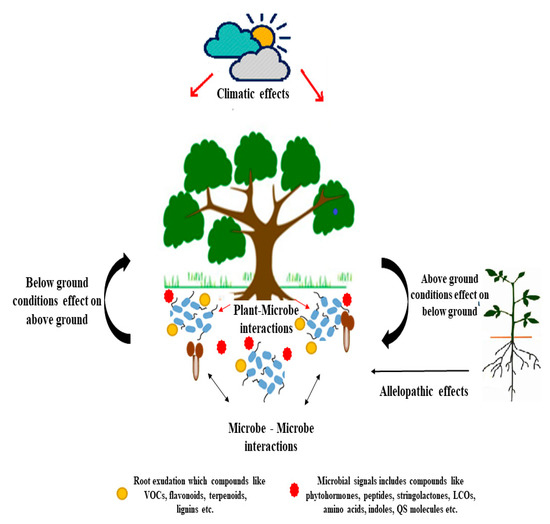
Figure 2. Schematic representation of phytomicrobiome on environment.
Plants mediate the immediate response using two types of receptors present on the outside and inside of the cell, which play an essential role in the recognition of a microbial reaction [62]. The outer receptor of the plant cell is governed by pattern recognition receptors (PRRs), a class of extracellular surface proteins. These receptors are evolutionary conserved and play a role in the recognition of the microbial cells. The activation of these receptors leads to intracellular signaling in plants and the biosynthesis of the molecules for that response. This response helps in the formation of a microbial biofilm in the rhizosphere as a result of the selective nutritional feeding of the beneficial microbial strains [63]. The composition of the root exudates helps the plant to be protected from pathogenic microbial strains [63,64]. The microbial colony created due to the plant PM is carefully maintained for a long time. These types of microbial interactions are particular to the plant root and shoot. These interactions form the channel of communication between the plant and microbes [65]. Many microbial species have been identified that secrete chemical compounds, extracellular enzymes, phytohormones, organic acids, and surface proteins based on the requirement of a particular instance. Examples of compounds recognized via the high-affinity cell surface PRRs of plants that activate an immune response are flagellins and lipopolysaccharides in Pseudomonas [66]. These signals control aspects of microbial as well as host plant activities and the overall community. The activation of the initial immune response of the plants towards the harmful pathogen also occurs by allowing access to beneficial endophytes [67].
Microbe-associated molecular patterns (MAMPs) also regulate immune responses, including antibiotic secretion. They have been shown to down-regulate during the secretion of plant root exudates in plant-associated Bacillus strains better to smooth the progress of root infection [68]. Plant-microbe beneficial interactions are very similar in the process as the pathogens infect the plant in different steps involving various chemical signals [69]. Plant signaling molecules mainly include the primary and secondary metabolites that belong to a variety of root exudates chemicals. Such signaling compounds are elevated in response to stress compounds, such as phytohormones, N-acyl-homoserine lactones (AHLs), phenols, and peptides, and are involved in microbe-to-microbe signaling as well as signaling between microbes and plants [68,69]. The Quorum sensing signaling molecules are produced by plant-associated bacteria and are utilized as signaling molecules for communication and for the regulation of gene expression [70]. AHLs have also been shown to affect the root development of Arabidopsis [71] and are involved in evoking induced systemic resistance (ISR), enabling plants to face various forms of biotic challenges which otherwise become lethal. Similarly, malic acid secretion was reported from Arabidopsis thaliana in response to foliage pathogenesis, which results in the promotion of biofilm formation in the rhizosphere of beneficial microbes [72]. Plants also face signaling materials produced by potential pathogens and responses by various chemical signals released by its activated defence and response systems [73]. Plants also exploit this microbial communication system in the modulation of gene expression in different microbial communities. The LuxR proteins of the bacteria activated by the plant signaling molecules [74]. This phenomenon is similar to the quorum sensing mechanism of the microbial strains. In the rhizospheric region, roots initiated the signaling between plant and microbes. The root produces the ethylene which acts as a signal with dual functions, perceiving biochemical cues and mediating the rhizospheric microbial assembly [75,76].
The root exudates of the plant are controlled by various factors such as time and space, physiology, nutritional status, the mechanical impendence of the plant, and nearby microbial activity in the rhizosphere [54,77,78]. The rhizosphere consists of three zones: the endorhizosphere, which includes the root tissue of the endodermis and cortical layers; the rhizoplane, which represents the root surface; and the ectorhizosphere, which is the soil near the root [79]. The microbial-rich nature of the rhizosphere in comparison to other soil was first proved by Hiltner in 1904 [80]. Similarly, the control of microbial load via root exudation was first predicted by Knudson in 1920 and Lyon and Wilson (1921) [81,82]. Root exudation utilizes most of the photosynthetically fixed carbon, and typically in the case of young seedlings it accounts for 30–40% of the total carbon fixation [83,84]. Root exudates contain majorly of carbon-based compounds along with ionic species, inorganic acids, oxygen, and water [84,85]. The organic component of exudate can be classified as low molecular weight compounds viz. amino acids, organic acids, sugars, phenolics; and an array of secondary metabolites and high high-molecular-weight compounds which comprise different proteins and mucilage compounds [86]. Some exudates are listed in Table 3.
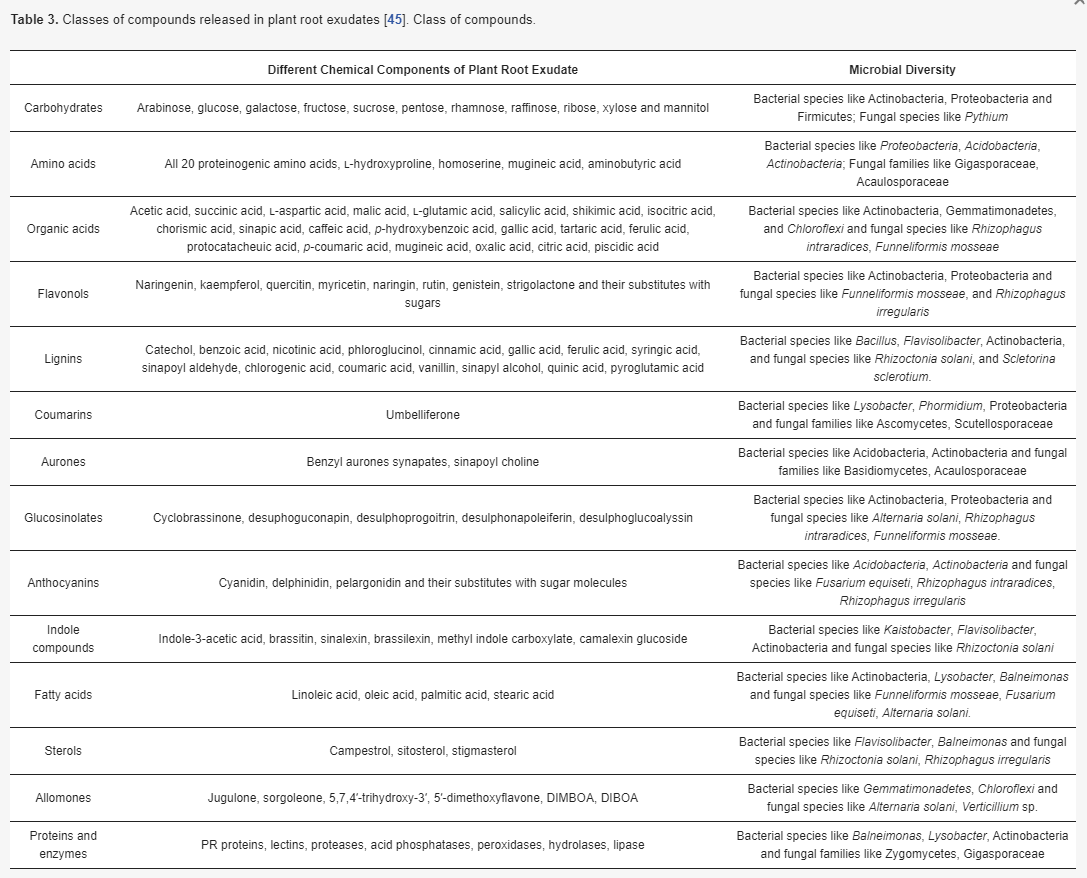
Table 3. Classes of compounds released in plant root exudates [45]. Class of compounds.
Plant root exudates are involved in interactions of the microbes with the rhizospheric region. The beneficial interactions of the plant root exudates are associated with the mycorrhizae, rhizobia, and soil bacteria involved in positive activities [86,87,88]. There are several reports available on the root exudates and their effect on the phytomicrobiome [89,90]. Rhizospheric PM inter communications represent the above-ground plant structure and microbial associations [73,90]; similarly, pathogen or herbivore attacks above ground can affect rhizospheric microbes. Injuries to the upper parts of plants have been shown to cause the stimulation of signaling compound production in plant roots [69,91]. The microbial community composition of the rhizosphere was observed to be altered in response to the increased photosynthetic rates under elevated CO2 conditions [92,93]. The involvement of high-throughput proteomics and metabolomics in understanding plant-microbe interactions and the signaling thereof can help in the development of effective, economic agricultural practices alleviating the fossil fuel-based crop inputs [94,95,96]. These studies also enable PM engineering for sustainable crop cultivation under presently variable climatic conditions [97,98].
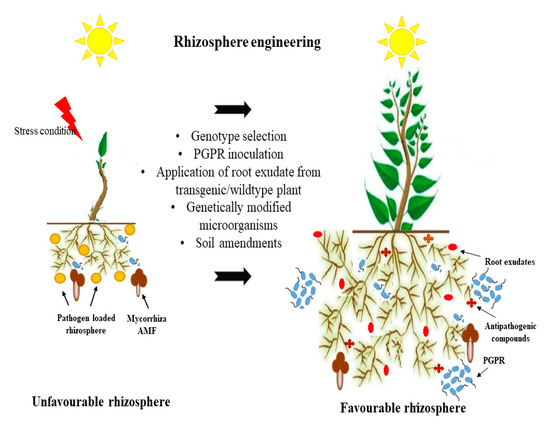
Figure 3. Rhizosphere engineering for improving plant growth promoting activities and balancing soil environment.
Plant engineers have developed genetically modified plants to cope with several problems related to soil biotic and abiotic stress. Engineered plants change their root exudation secretion and the pH of the rhizosphere. This change stimulates a beneficial shift in microorganism activity. This change in the rhizosphere stimulates a beneficial shift in microorganism activity. Apart from plant engineering, microbial engineering plays an essential role in agriculture research, especially with PGPR, biological nitrogen fixation, the solubilization of phosphorous and iron, and the modulation of phytohormone and biocontrol activity [102]. Due to the multiple interactive biomes of the rhizosphere, a change in these interactions has the potential to alter plant productivity, health, and soil properties. Therefore, rhizosphere engineering is promising for soil improvement, crop quality, and productivity. Several studies have promoted rhizosphere engineering. In this series, extensive studies have been carried out on the plant and microbe interaction. For instance, specific Pseudomonas spp. of the maize rhizosphere influence the lateral root and root hair for plant growth [103], and Phyllobacterium brassicacearum stimulates the ethylene pathway for the development of root hairs [104]. Classical breeding approaches have developed resistance to significant pathogens, such as Pseudomonas syringae, and the Xanthomonas species. The engineering of PGPR microbes involves the introgression of the chromosome from resistant to susceptible lines, such as substituting the chromosome of the susceptible wheat line S-615 and rescuing from the resistant wheat line Apex chromosome 5B, which results in chromosome substituent lines SA5B that are as resistant as Apex against common rot [103,104,105]. It was noticed that plant density might be playing a role in the existence of the phytomicrobiome. Traditional breeding considers the plant densities as an essential part of crop improvement. Genetic potential for yield and other traits is fully expressed when plants are grown at very low plant densities, and it is significantly suppressed at high plant densities [106]. Studies on the engineering of the plant-microbe interaction for sustainable agriculture are listed in Table 4.
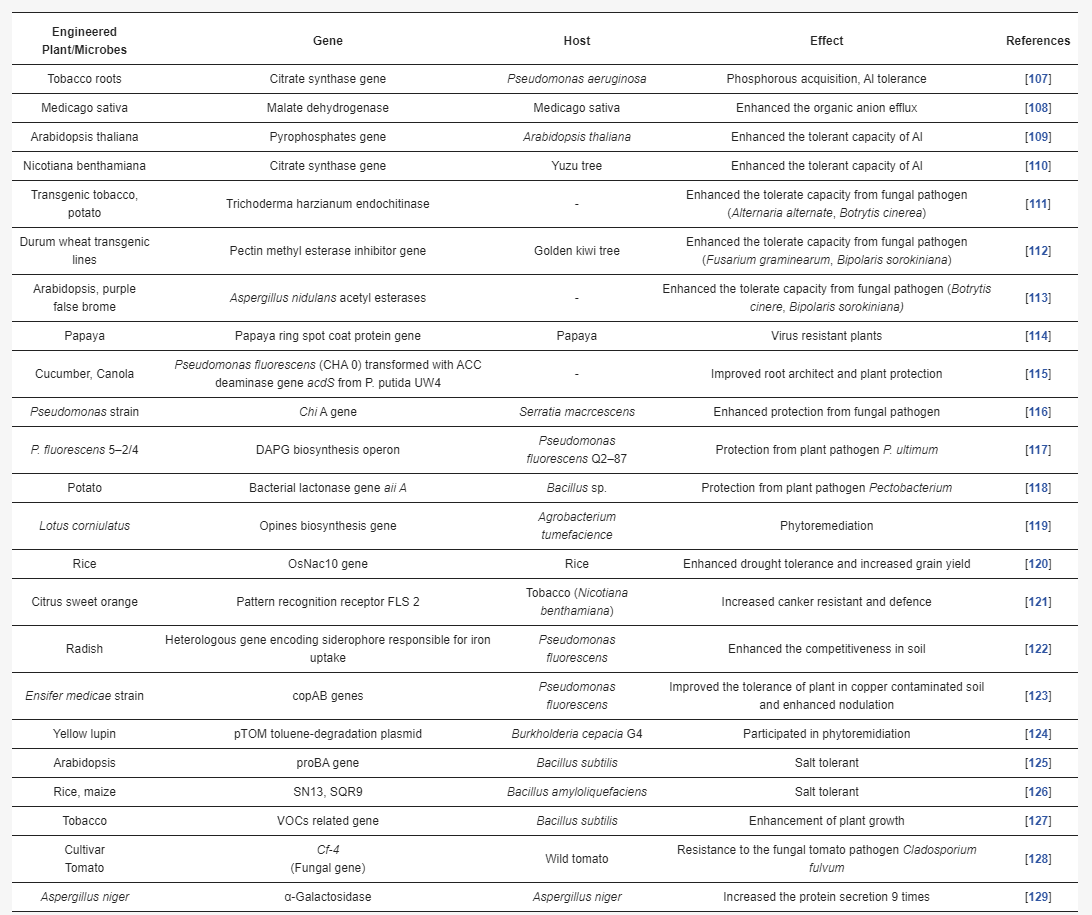
Table 4. Phytomicrobiome engineering: engineered plant/microbes for rhizosphere enrichment.
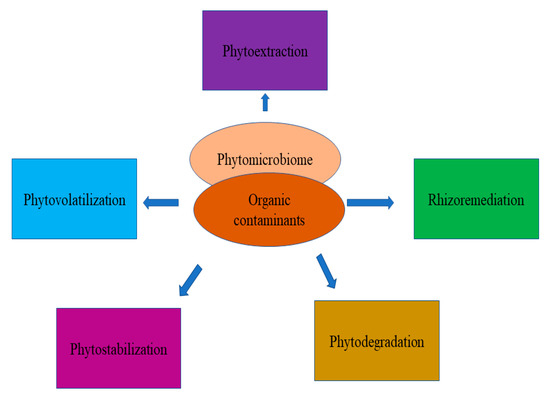
Figure 4. Overview of the phytomicrobiome in the degradation of organic pollutants from soil.
Thus, potential investigations of the PM can support rhizoremediation technology for the better management of soil contaminants and the rejuvenation of various agricultural lands which are affected due to industrial activities. Martínez et al. [144] reported on the fungal community assembly of holm oak trees (Quercus ilex) growing in soil contaminated with trace elements after metalliferous tailing spills. It was concluded that the fungal community composition in ectomycorrhiza affects the plant performance in metal-contaminated soils, thus controlling the phytoremediation efficiency [46,141]. Such studies reflect the necessity of exploration of the PM to understand the critical role of the microbial community in managing the plant traits that can be utilized in purposes like rhizoremediation. Plants and associated microbial communities play an essential role in a constructed wetland [145,146,147]. The endophytic bacterial communities of Juncus acutus are able to degrade the pollutant from the soil of wetlands [44]. Phytoremediation capabilities can be modified by bioaugmentation methods, enhancing the accumulation of soil contaminants in plant tissues, thus sustaining the plant growth and development, along with the cleanup of soil pollutants [148,149,150]. PM engineering could improve plant survival in contaminated soil by planting willow plants. Petroleum-contaminated soils are treated with gamma irradiation and different inoculation strategies [42]. With a better understanding of the synergistic relationships of PM, rhizoremediation can be developed as a promising technology to tackle the problem of soils contaminated with a wide variety of contaminants [151,152,153]. It is now a well-established fact that the rhizoremediation of contaminated soil depends on PM functioning, and the more we understand the relationships of the PM, the better we can utilize the PM for rhizoremediation. This not only enables land reclamation but also the better utilization of various plant species for rhizoremediation, as well as the sustainable cultivation of multiple crops without harming the natural conditions.
* Authors to whom correspondence should be addressed.
† Both authors contributed equally to this manuscript.
Abstract
Recent crop production studies have aimed at an increase in the biotic and abiotic tolerance of plant communities, along with increased nutrient availability and crop yields. This can be achieved in various ways, but one of the emerging approaches is to understand the phytomicrobiome structure and associated chemical communications. The phytomicrobiome was characterized with the advent of high-throughput techniques. Its composition and chemical signaling phenomena have been revealed, leading the way for “rhizosphere engineering”. In addition to the above, phytomicrobiome studies have paved the way to best tackling soil contamination with various anthropogenic activities. Agricultural lands have been found to be unbalanced for crop production. Due to the intense application of agricultural chemicals such as herbicides, fungicides, insecticides, fertilizers, etc., which can only be rejuvenated efficiently through detailed studies on the phytomicrobiome component, the phytomicrobiome has recently emerged as a primary plant trait that affects crop production. The phytomicrobiome also acts as an essential modifying factor in plant root exudation and vice versa, resulting in better plant health and crop yield both in terms of quantity and quality. Not only supporting better plant growth, phytomicrobiome members are involved in the degradation of toxic materials, alleviating the stress conditions that adversely affect plant development. Thus, the present review compiles the progress in understanding phytomicrobiome relationships and their application in achieving the goal of sustainable agriculture.Keywords: phytomicrobiome; rhizosphere engineering; organic contamination; microorganisms
1. Introduction
The phytomicrobiome (PM) can be described as the microbial community associated with a plant that constitutes the whole root as well as the shoot parts. We know that the advent of photosynthesis changed the overall evolutionary fate on earth [1]. On the earth, at present, plants are one of the main entry sources of energy [2]. Therefore, non-photosynthetic organisms including humans remain reliant on plants for their energy needs. Similarly, microorganisms form a complex association with plant parts, so this photo community system, which includes plants and microbes, is comprised of regulated molecular signals that modulate the activity of each other and satisfy mutual benefits [3]. The awareness of plant-associated microbial communities started with research on legume–microbial associations, where an intricate signaling system between the plant and microorganisms in the rhizosphere was found [4]. The rhizospheric region of the plant contains the pool of microbial strains acting as plant growth-promoting rhizobacteria (PGPR). Further research related to deciphering molecular signals and biochemical compounds, which take place in the rhizosphere, has revealed a sophisticated and complex signaling mechanism that helps plants to thrive in their surrounding environment (Table 1).
Table 1. Signaling and chemical interactions of the rhizosphere.
The microbial community interacts with the plant not only in the rhizospheric region but also in the phyllosphere, which contains a wide array of living organisms such as bacteria, fungi, yeasts, algae, nematodes, etc. It influences the phytophysiology and overall development [18]. The biotic and abiotic conditions regulate the microbiome composition of the phyllosphere [19]. This PM also varies with biotic factors, such as a pathogenic organism causing disease, where the host plant microbiome varies with disease progression [20]. Thus, understanding the phyllosphere microbial community gives an informative overview of plant health. PM analysis requires a holistic study involving the different techniques of genomics, transcriptomics, proteomics, and metabolomics to determine the relationship between the plant and its associated microbial community. An amplified rDNA restriction analysis (ARDRA) is usually utilized for characterizing the bacterial community present in any given sample, which in short involves enzymatic amplification followed by the restriction digestion of 16S rRNA amplified fragments [21]. Advanced techniques like Illumina-based microbiome analysis further helps to analyze the microbial community and their dynamic relationships in much less time [20,22]. Thus, recently, much research has been carried out concerning the PM structure and composition and their variations under different plant conditions.
2. Phytomicrobiome: Its Composition and Biomass
Plants are surrounded by various microbial communities which affect them either positively or negatively. These communities are present around all plant parts, including the roots and shoots, of which the most studied are the below-ground or root-associated microbial communities [23]. The rhizomicrobiome constitutes of root-associated microbes, the phyllomicrobiome constitutes of shoot-associated microbial communities, and the endomicrobiome consists of microbial communities present in the inner plant tissue system. Many studies have investigated the different components of the PM, revealing the wide diversity of microbes which varies from crop to crop [24]. It varies with the developmental stage of the same crop, as well as with the particular developmental stage in the presence of stress conditions [25]. A study with common bean, soybean, and canola related to the effect of biotic and abiotic stress on the phyllosphere composition at three locations in Ontario, Canada, revealed that the phyllosphere community structure changes under seasonal variations and that these community variations are very active [18]. Similarly, da Silva et al. (2014) carried out a study on the endomicrobiomes of different transgenic and nontransgenic maize genotypes. They reported about different factors which controlled the endomicrobiome microbial assemblage and identified these factors’ proteins which were expressed in the transgenic genotypes [26] (Figure 1).
Figure 1. Factors regulating the microbiome composition.
Studies related to PM compositions have revealed that plants are in dynamic relationships with the surrounding microbial communities, which show variations as per the plant developmental stage and their environmental conditions. Either the rhizosphere or phyllosphere or even the endophytic microbes are seen to enable plant establishment in their surroundings. It is very interesting to note that the same plant grown in different soils shows the recruitment of the same microbial communities for their establishment, which strongly reflects the modifications of the soil microbiota by plants. This underlies an intricate pathway of signaling compounds for community assembly, which opens a new avenue of research. The root microbiome of the plant of different species shows variations in community structure, along with the presence of Actinobacteria, Proteobacteria, and Bacteriodetes as conserved phyla in the rhizomicrobiome of these plant species [27]. The plants Arabidopsis thaliana, A. lyrata, A. helleri, and Cardamine hirsute, which were grown in the same soil in the mentioned study, reflect the requirement for the exploration of PM signaling for sustainable cultivation.
Similarly, in phyllosphere studies it has been observed by many workers that there is little variation in the phyllomicrobiome among the similar plant species, even those planted in various geographical areas [28,29]. Recently, these PM-associated microorganisms were found to be crucial in imparting tolerance against biotic and abiotic stress. The role of the rhizosphere communities is well established, but in recent years workers have reported the involvement of various phyllospheric communities in plant stress alleviation. The phyllosphere bacteria of the Bacillus genus have been reported to mitigate drought stress in rice by previous researchers [30]. The phyllosphere isolates were found to be involved in the activation of the plant antioxidant system by an increase in proline-like osmolytes; phenolic content; and the activity of antioxidative enzymes, such as superoxide dismutase, catalase, peroxidase, etc. [31]. Such studies are now easy to conduct with the advent of what we call “next-generation sequencing”, which includes the methods of 454 pyrosequencing, Illumina sequencing, etc. As compared to the traditional techniques, these tools are advantageous and can be used for multidirectional analysis. Further, these techniques are supplemented by metaproteomic analysis, revealing the complex networks of these PM [32]. Thus, we can say that understanding the ecology of the PM is presently an important thrust area of research to achieve sustainability in agriculture in the coming years. Some of the recent PM studies are presented in Table 2.

Table 2. Phytomicrobiome ecology: a recent overview of the techniques involved and the communities identified.
3. Phytomicrobiome Signaling
Signaling molecules play a crucial role in communication among plants and the microbe [45]. The legume-rhizobium system is one of the widely studied systems for plant-microbe interactions, and many chemical signals are identified from this system [45,46]. The mycorrhizae group of fungi also has similar interactions [47]. The plant partner releases the chemical signals (flavonoid) in the form of root exudates, which makes a suitable condition for the synthesis of the signaling molecules lipo-chitooligosaccharides (LCOs) in microbial strains [48]. The LCOs consist of receptors that have kinase activity and are involved in root hormone profile variations and root nodule development [49,50]. The PM community composition varies as per the signaling by plant parts in response to the environmental conditions around plant [51,52,53,54,55]. The LCOs are found to be involved in plant growth stimulation [55,56,57], the elaboration of the root system [58], the acceleration of flowering and increased fruit yield [59], and stimulating early somatic embryo development [4,50,57,60]. The enhanced germination and growth of seedlings, along with the LCOs’ positive effect on mitotic cell division, suggest an activated meristem activity. Recently, various products based on LCOs have been applied to several million ha of cropland, mainly corn and soybean, each year during seed sowing. The LCO effects are much higher under the presence of stress (salt, drought, cold) as compared to optimum conditions [61] (Figure 2).
Figure 2. Schematic representation of phytomicrobiome on environment.
Plants mediate the immediate response using two types of receptors present on the outside and inside of the cell, which play an essential role in the recognition of a microbial reaction [62]. The outer receptor of the plant cell is governed by pattern recognition receptors (PRRs), a class of extracellular surface proteins. These receptors are evolutionary conserved and play a role in the recognition of the microbial cells. The activation of these receptors leads to intracellular signaling in plants and the biosynthesis of the molecules for that response. This response helps in the formation of a microbial biofilm in the rhizosphere as a result of the selective nutritional feeding of the beneficial microbial strains [63]. The composition of the root exudates helps the plant to be protected from pathogenic microbial strains [63,64]. The microbial colony created due to the plant PM is carefully maintained for a long time. These types of microbial interactions are particular to the plant root and shoot. These interactions form the channel of communication between the plant and microbes [65]. Many microbial species have been identified that secrete chemical compounds, extracellular enzymes, phytohormones, organic acids, and surface proteins based on the requirement of a particular instance. Examples of compounds recognized via the high-affinity cell surface PRRs of plants that activate an immune response are flagellins and lipopolysaccharides in Pseudomonas [66]. These signals control aspects of microbial as well as host plant activities and the overall community. The activation of the initial immune response of the plants towards the harmful pathogen also occurs by allowing access to beneficial endophytes [67].
Microbe-associated molecular patterns (MAMPs) also regulate immune responses, including antibiotic secretion. They have been shown to down-regulate during the secretion of plant root exudates in plant-associated Bacillus strains better to smooth the progress of root infection [68]. Plant-microbe beneficial interactions are very similar in the process as the pathogens infect the plant in different steps involving various chemical signals [69]. Plant signaling molecules mainly include the primary and secondary metabolites that belong to a variety of root exudates chemicals. Such signaling compounds are elevated in response to stress compounds, such as phytohormones, N-acyl-homoserine lactones (AHLs), phenols, and peptides, and are involved in microbe-to-microbe signaling as well as signaling between microbes and plants [68,69]. The Quorum sensing signaling molecules are produced by plant-associated bacteria and are utilized as signaling molecules for communication and for the regulation of gene expression [70]. AHLs have also been shown to affect the root development of Arabidopsis [71] and are involved in evoking induced systemic resistance (ISR), enabling plants to face various forms of biotic challenges which otherwise become lethal. Similarly, malic acid secretion was reported from Arabidopsis thaliana in response to foliage pathogenesis, which results in the promotion of biofilm formation in the rhizosphere of beneficial microbes [72]. Plants also face signaling materials produced by potential pathogens and responses by various chemical signals released by its activated defence and response systems [73]. Plants also exploit this microbial communication system in the modulation of gene expression in different microbial communities. The LuxR proteins of the bacteria activated by the plant signaling molecules [74]. This phenomenon is similar to the quorum sensing mechanism of the microbial strains. In the rhizospheric region, roots initiated the signaling between plant and microbes. The root produces the ethylene which acts as a signal with dual functions, perceiving biochemical cues and mediating the rhizospheric microbial assembly [75,76].
The root exudates of the plant are controlled by various factors such as time and space, physiology, nutritional status, the mechanical impendence of the plant, and nearby microbial activity in the rhizosphere [54,77,78]. The rhizosphere consists of three zones: the endorhizosphere, which includes the root tissue of the endodermis and cortical layers; the rhizoplane, which represents the root surface; and the ectorhizosphere, which is the soil near the root [79]. The microbial-rich nature of the rhizosphere in comparison to other soil was first proved by Hiltner in 1904 [80]. Similarly, the control of microbial load via root exudation was first predicted by Knudson in 1920 and Lyon and Wilson (1921) [81,82]. Root exudation utilizes most of the photosynthetically fixed carbon, and typically in the case of young seedlings it accounts for 30–40% of the total carbon fixation [83,84]. Root exudates contain majorly of carbon-based compounds along with ionic species, inorganic acids, oxygen, and water [84,85]. The organic component of exudate can be classified as low molecular weight compounds viz. amino acids, organic acids, sugars, phenolics; and an array of secondary metabolites and high high-molecular-weight compounds which comprise different proteins and mucilage compounds [86]. Some exudates are listed in Table 3.

Table 3. Classes of compounds released in plant root exudates [45]. Class of compounds.
Plant root exudates are involved in interactions of the microbes with the rhizospheric region. The beneficial interactions of the plant root exudates are associated with the mycorrhizae, rhizobia, and soil bacteria involved in positive activities [86,87,88]. There are several reports available on the root exudates and their effect on the phytomicrobiome [89,90]. Rhizospheric PM inter communications represent the above-ground plant structure and microbial associations [73,90]; similarly, pathogen or herbivore attacks above ground can affect rhizospheric microbes. Injuries to the upper parts of plants have been shown to cause the stimulation of signaling compound production in plant roots [69,91]. The microbial community composition of the rhizosphere was observed to be altered in response to the increased photosynthetic rates under elevated CO2 conditions [92,93]. The involvement of high-throughput proteomics and metabolomics in understanding plant-microbe interactions and the signaling thereof can help in the development of effective, economic agricultural practices alleviating the fossil fuel-based crop inputs [94,95,96]. These studies also enable PM engineering for sustainable crop cultivation under presently variable climatic conditions [97,98].
4. Rhizosphere Engineering
Rhizosphere engineering can boost plant health and alter the activity of root-associated microorganisms for the development of sustainable agriculture. The phytomicrobiome belongs to the genus Rhizobia, Pseudomonas, Bacillus, Burkholderia, Azospirillum, Klebsiella, and Gluconacetobacter applied in agricultural purposes [99]. To utilize the potential of the rhizosphere for the development of plants and the associated environment, it is essential to acknowledge the different root exudate molecules and their interactions with the rhizosphere microflora. The understanding of rhizosphere interactions is essential for the creation of sustainable agroecosystems [100]. The manipulation of the plant and its associated microorganisms changes the rhizosphere through the release of root exudation compounds, which positively influence microbial signaling compounds. Root exudates vary with the plant genotype and species. On the other side, microorganisms secrete signaling compounds categorized into phytohormones, extracellular enzymes, organic acids, antibiotics, volatile signals, and quorum-sensing molecules [101]. Several studies have investigated the modification of plant and rhizosphere microflora for the enrichment of the rhizospheric zone for sustainable agriculture (Figure 3).
Figure 3. Rhizosphere engineering for improving plant growth promoting activities and balancing soil environment.
Plant engineers have developed genetically modified plants to cope with several problems related to soil biotic and abiotic stress. Engineered plants change their root exudation secretion and the pH of the rhizosphere. This change stimulates a beneficial shift in microorganism activity. This change in the rhizosphere stimulates a beneficial shift in microorganism activity. Apart from plant engineering, microbial engineering plays an essential role in agriculture research, especially with PGPR, biological nitrogen fixation, the solubilization of phosphorous and iron, and the modulation of phytohormone and biocontrol activity [102]. Due to the multiple interactive biomes of the rhizosphere, a change in these interactions has the potential to alter plant productivity, health, and soil properties. Therefore, rhizosphere engineering is promising for soil improvement, crop quality, and productivity. Several studies have promoted rhizosphere engineering. In this series, extensive studies have been carried out on the plant and microbe interaction. For instance, specific Pseudomonas spp. of the maize rhizosphere influence the lateral root and root hair for plant growth [103], and Phyllobacterium brassicacearum stimulates the ethylene pathway for the development of root hairs [104]. Classical breeding approaches have developed resistance to significant pathogens, such as Pseudomonas syringae, and the Xanthomonas species. The engineering of PGPR microbes involves the introgression of the chromosome from resistant to susceptible lines, such as substituting the chromosome of the susceptible wheat line S-615 and rescuing from the resistant wheat line Apex chromosome 5B, which results in chromosome substituent lines SA5B that are as resistant as Apex against common rot [103,104,105]. It was noticed that plant density might be playing a role in the existence of the phytomicrobiome. Traditional breeding considers the plant densities as an essential part of crop improvement. Genetic potential for yield and other traits is fully expressed when plants are grown at very low plant densities, and it is significantly suppressed at high plant densities [106]. Studies on the engineering of the plant-microbe interaction for sustainable agriculture are listed in Table 4.

Table 4. Phytomicrobiome engineering: engineered plant/microbes for rhizosphere enrichment.
5. Phytomicrobiome and Rhizoremediation
Microbial strains have the potential to degrade organic pollutants from the soil and the rhizospheric region [130,131,132]. These organic pollutants are mainly pesticides, antibiotics, and steroids. Through their metabolism, they can able to degrade toxic man-made chemicals [132,133,134,135]. The rhizospheric region in plants consists of a pool of specific strains that facilitate the ability to degrade toxic pollutants, as well as promote plant growth [136,137,138]. The plant-associated mechanisms involved in the bioremediation using plants are known as phytotransformation, phytoextraction, phytostabilization, and phytovolatilization [139]. The technique is considered “Green Technology”, as it involves better management of soil contaminants along with the surrounding water and air. However, the success of this technology is limited by many constraints, especially the adaptation of the plant species to contaminated soils. The rhizospheric microbes can use organic contaminant as the sole source of nutrients [140]. These strains interacted with plants in a symbiotic manner and reduce a load of pollutants [141]. Rhizoremediation (RR) is related to this and involves the degradation of soil pollutants through the rhizospheric plant-microbe interactions. We know that plant development and its establishment in a particular habitat depends upon a physiologically robust root system. Recently, it had been found that plant roots not only establish the plant in the soil physically but also modify the soil conditions through their exudation profiles, recruiting various microbial species to help the plant accommodate to the habitat and develop better [142]. Thus, the rhizomicrobiome characteristics and composition can be helpful in the prediction of the rhizoremediation efficiency of plants. The rhizomicrobiome community forms a mega consortium under the guidance of the root exudation signaling of plants for sustaining plant growth and development, apart from the modification of physical soil conditions, as well as ecology. The plant microbiome has been shown to have a crucial role to play in nutrient acquisition, disease resistance, and stress tolerance [143]. Contaminated soil represents an ideal stress condition under which the plant undergoes multiple varieties of dynamic plant-microbe signaling interactions to combat unfavorable conditions and transform the condition to better suit the relationship between the plant metabolism and the PM (Figure 4).
Figure 4. Overview of the phytomicrobiome in the degradation of organic pollutants from soil.
Thus, potential investigations of the PM can support rhizoremediation technology for the better management of soil contaminants and the rejuvenation of various agricultural lands which are affected due to industrial activities. Martínez et al. [144] reported on the fungal community assembly of holm oak trees (Quercus ilex) growing in soil contaminated with trace elements after metalliferous tailing spills. It was concluded that the fungal community composition in ectomycorrhiza affects the plant performance in metal-contaminated soils, thus controlling the phytoremediation efficiency [46,141]. Such studies reflect the necessity of exploration of the PM to understand the critical role of the microbial community in managing the plant traits that can be utilized in purposes like rhizoremediation. Plants and associated microbial communities play an essential role in a constructed wetland [145,146,147]. The endophytic bacterial communities of Juncus acutus are able to degrade the pollutant from the soil of wetlands [44]. Phytoremediation capabilities can be modified by bioaugmentation methods, enhancing the accumulation of soil contaminants in plant tissues, thus sustaining the plant growth and development, along with the cleanup of soil pollutants [148,149,150]. PM engineering could improve plant survival in contaminated soil by planting willow plants. Petroleum-contaminated soils are treated with gamma irradiation and different inoculation strategies [42]. With a better understanding of the synergistic relationships of PM, rhizoremediation can be developed as a promising technology to tackle the problem of soils contaminated with a wide variety of contaminants [151,152,153]. It is now a well-established fact that the rhizoremediation of contaminated soil depends on PM functioning, and the more we understand the relationships of the PM, the better we can utilize the PM for rhizoremediation. This not only enables land reclamation but also the better utilization of various plant species for rhizoremediation, as well as the sustainable cultivation of multiple crops without harming the natural conditions.





